Overview
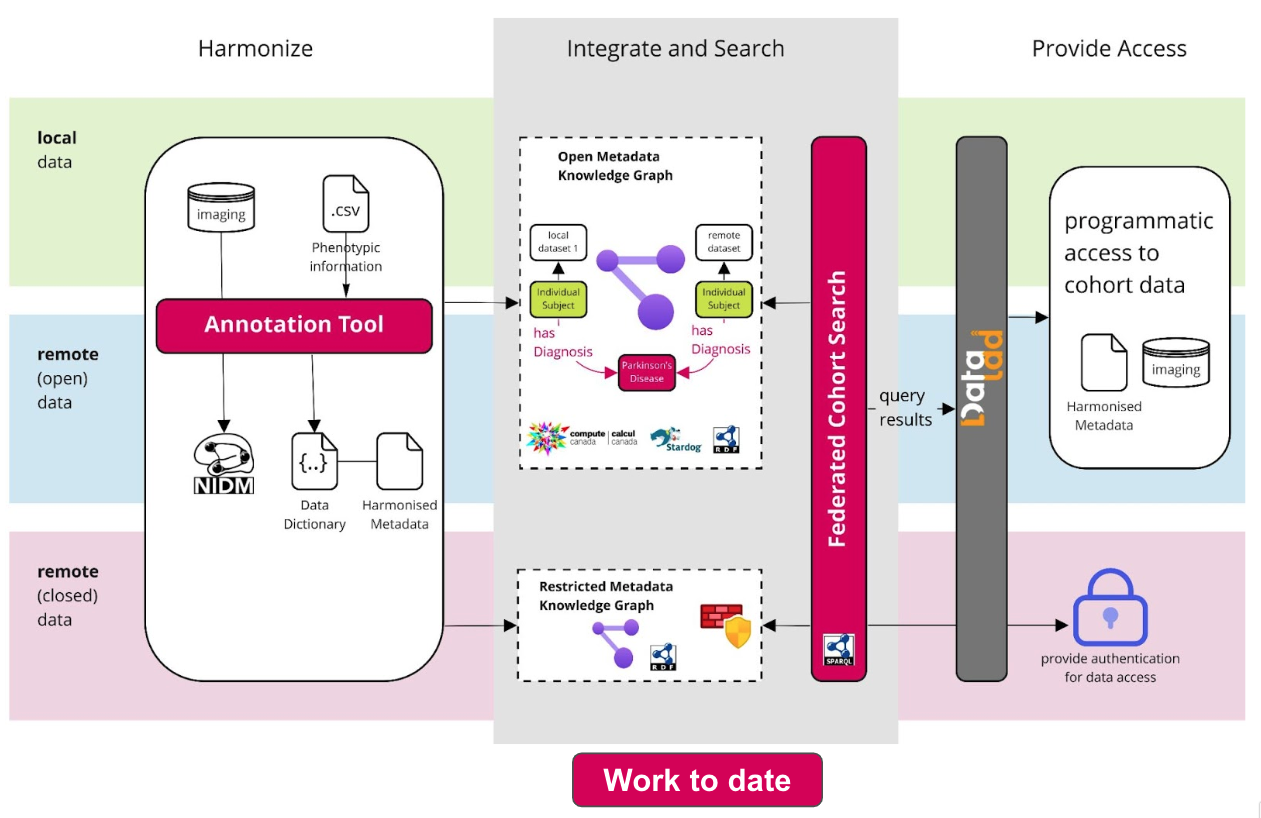
The goal of Neurobagel is to make describing and finding neuroimaging
data easy. Searching across data that live on distributed storage systems
is tricky, and the technologies that make it possible are not usually
something researchers have experience with or time to learn.
We are building web-based, graphical user interfaces that make these steps
easier:
- 1. Annotate and harmonize phenotypic data
- 2. Define and find subject-level cohorts across neuroimaging datasets
These tools are still in early development, but we are excited to share
them with the neuroimaging community and are always looking for feedback.
Take a look at our github
repositories and our roadmap below to get a sense of what we are
doing at the moment!